docs/learning.qmd
participant_details <- here("data-raw/dime/participant_details.csv") |>
import_dime()pivot_longer() and pivot_wider() functions from the tidyr package to pivot data.Time: ~6 minutes.
A very effective way to learn is to recall and describe to someone else what you’ve learned. So before we continue this session, take some time to think about what you’ve learned in the last two days.
Briefly walk through it again after they’ve read it and explain it a bit more, making use of the tables and graphs. Pivoting is often a difficult concept to grasp, to it’s good to go over it a few times. Emphasise the difficulty of it, to remind them it is ok to not get it.
Time: ~6 minutes.
Rarely is our data in the exact format we need or want it to be in to effectively process, analyse, or visualize it. Often we’ll need to do some heavy reshaping. One very powerful way to reshape data is with “pivoting”. Pivoting is when you take a dataset’s rows and convert them to columns or convert columns to rows, and in many different combinations of this.
The tidyr package within the tidyverse contains two wonderful functions for pivoting: pivot_longer() and pivot_wider(). There is a well written documentation on pivoting in the tidyr website that can explain more about it that you can read at another time.
Most use cases of pivoting are to convert to the longer form, which you can do with pivot_longer() and is what we will cover first in this session. There are many reasons pivoting longer is the most common pivot direction.
An example is if you ever need to enter data. Entering data, especially in a spreadsheet tool, in the wide form is much easier and more time efficient than entering data in long form. Consider the scenario where you are entering data from a participant for glucose values that were measured every 30 minutes. So you might enter data like this:
| person_id | glucose_0 | glucose_30 | glucose_60 |
|---|---|---|---|
| 1 | 5.6 | 7.8 | 4.5 |
| 2 | 4.7 | 9.5 | 5.3 |
| 3 | 5.1 | 10.2 | 4.2 |
If working with a spreadsheet, is this is to use tab to move to the next cell and continue entering the data. However, when it comes time to analyze the data, this wide form is very inefficient and difficult to computationally and statistically work with. So, we do data entry in wide and use functions like pivot_longer() to get the data ready for analysis. See Figure 11.1 below that visually shows what happens when you pivot from wide to long.
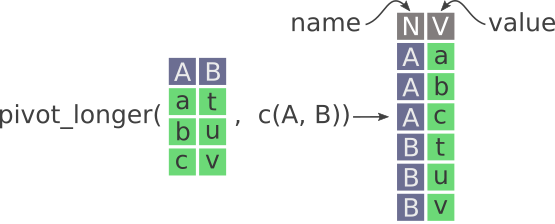
name and value. Notice how the values in A and B columns are stacked on top of each other in the newly created V column.
If you had, for instance, an ID column for each participant, the pivoting would look like what is shown in Figure 11.2.
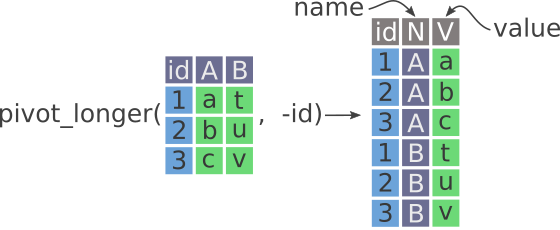
id column. New columns are called name and value, as well as the old id column. Notice how, unlike the previous image, the id column is excluded when pivoting into the data on the right.
Pivoting is a conceptually challenging thing to grasp, so don’t be discouraged if you can’t understand how it works yet or of the ways you could use. As you practice using it, you will slowly begin to understand it. With pivot_longer(), the first argument is the data itself. The other arguments are:
cols: The columns to use to convert to long form. The input is a vector made using c() that contains the column names, like you would use in select() (e.g. you can use the select_helpers like starts_with(), or - minus to exclude).names_to: Optional, the default is name. If provided, it will be the name of the newly created column (as a quoted character) that contains the original column names.values_to: Optional, the default is value. Like names_to, sets the name of the new columns.Both pivot_longer() and its opposite pivot_wider(), which we will cover later in the session, are incredibly powerful functions. We can’t show close to everything it can do in this workshop, but if you want to learn more, read up on the documentation for it.
When you’re ready to continue, place the sticky/paper hat on your computer to indicate this to the teacher 👒 🎩
Emphasise that there are a lot of really powerful ways you can use pivot_longer() and to point out looking to the documentation to see some examples of that.
Our CGM data is already in a long format, but we eventually want to join it with the participant details data. The participant details data is in a wide format. Let’s start with importing it in the setup code chunk of our docs/learning.qmd file:
docs/learning.qmd
participant_details <- here("data-raw/dime/participant_details.csv") |>
import_dime()Run the code in the setup code chunk by running each line with Ctrl-EnterCtrl-Enter. Then, let’s move to the end of our docs/learning.qmd file and create a new header called ## Pivot longer. Below that, create a new code chunk with Ctrl-Alt-ICtrl-Alt-I or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “new chunk”). We’ll start with seeing what the data looks like:
docs/learning.qmd
participant_details# A tibble: 60 × 6
id gender age_years intervention start_date end_date
<dbl> <chr> <dbl> <chr> <date> <date>
1 101 woman 45.6 baseline 2021-03-15 2021-03-22
2 101 woman 45.6 low bioactive diet 2021-03-23 2021-04-06
3 101 woman 45.6 high bioactive diet 2021-05-11 2021-05-25
4 102 man 43.7 baseline 2021-03-15 2021-03-22
5 102 man 43.7 high bioactive diet 2021-03-23 2021-04-06
6 102 man 43.7 low bioactive diet 2021-05-11 2021-05-25
7 104 man 37.3 baseline 2021-06-21 2021-06-28
8 104 man 37.3 low bioactive diet 2021-06-29 2021-07-13
9 104 man 37.3 high bioactive diet 2021-08-23 2021-09-06
10 105 man 35.8 baseline 2021-04-13 2021-04-21
# ℹ 50 more rowsYou can see that the data, while the data has the intervention column in the long format, the dates are in the wide format, with start_date and end_date. Our CGM and sleep only has a single date column, so we can’t yet join these datasets with participant_details. We first need to pivot the data to the long format. Let’s add a pipe to participant_details to output into pivot_longer():
docs/learning.qmd
participant_details |>
pivot_longer()Error in `pivot_longer()`:
! `cols` must select at least one column.This give an error because we haven’t told it which columns to use yet. We want to pivot only the date columns. We can do this using tidyselect helpers, for instance the helper function ends_with() is perfect for this case, since both date columns end with date.
docs/learning.qmd
participant_details |>
pivot_longer(ends_with("date"))# A tibble: 120 × 6
id gender age_years intervention name value
<dbl> <chr> <dbl> <chr> <chr> <date>
1 101 woman 45.6 baseline start_date 2021-03-15
2 101 woman 45.6 baseline end_date 2021-03-22
3 101 woman 45.6 low bioactive diet start_date 2021-03-23
4 101 woman 45.6 low bioactive diet end_date 2021-04-06
5 101 woman 45.6 high bioactive diet start_date 2021-05-11
6 101 woman 45.6 high bioactive diet end_date 2021-05-25
7 102 man 43.7 baseline start_date 2021-03-15
8 102 man 43.7 baseline end_date 2021-03-22
9 102 man 43.7 high bioactive diet start_date 2021-03-23
10 102 man 43.7 high bioactive diet end_date 2021-04-06
# ℹ 110 more rowsAwesome! We now have a long format with dates in a single column. But we also have new column names that are a bit ambiguous, like name and value. We can tell pivot_longer() to give them more descriptive names with names_to and values_to. But, we also don’t need the name column, since implicitly we know that the last date is the end date and the first date is the start date. So, we can set names_to = NULL to drop the new name column from being created.
docs/learning.qmd
participant_details |>
pivot_longer(ends_with("date"), names_to = NULL, values_to = "date")# A tibble: 120 × 5
id gender age_years intervention date
<dbl> <chr> <dbl> <chr> <date>
1 101 woman 45.6 baseline 2021-03-15
2 101 woman 45.6 baseline 2021-03-22
3 101 woman 45.6 low bioactive diet 2021-03-23
4 101 woman 45.6 low bioactive diet 2021-04-06
5 101 woman 45.6 high bioactive diet 2021-05-11
6 101 woman 45.6 high bioactive diet 2021-05-25
7 102 man 43.7 baseline 2021-03-15
8 102 man 43.7 baseline 2021-03-22
9 102 man 43.7 high bioactive diet 2021-03-23
10 102 man 43.7 high bioactive diet 2021-04-06
# ℹ 110 more rowsGreat! But, maybe you may have noticed something else here. If we want to join this data with the CGM data, we need to have a single row for each day of the study that the participant has data for. Right now, the participant_details data has only the date for the first day and for the last day. So if we join the CGM or sleep data, we would only get the data for those two dates. What we want is to have all the days included between the start and end date for each participant. The tidyr package has many wonderful functions that help tidy up data, including a function for this particular situation! The function is called complete() because it will fill in data where you want a sequence with the missing dates in that sequence. For our situation, that is filling in all the missing dates between the start and end date for each participant.
Since we want to complete the data for each participant for each intervention, we will need to group the data. We could select each column to group by, but we could also use the pick() function we used before to select all the columns except for the date column.
docs/learning.qmd
# A tibble: 120 × 5
id gender age_years intervention date
<dbl> <chr> <dbl> <chr> <date>
1 101 woman 45.6 baseline 2021-03-15
2 101 woman 45.6 baseline 2021-03-22
3 101 woman 45.6 low bioactive diet 2021-03-23
4 101 woman 45.6 low bioactive diet 2021-04-06
5 101 woman 45.6 high bioactive diet 2021-05-11
6 101 woman 45.6 high bioactive diet 2021-05-25
7 102 man 43.7 baseline 2021-03-15
8 102 man 43.7 baseline 2021-03-22
9 102 man 43.7 high bioactive diet 2021-03-23
10 102 man 43.7 high bioactive diet 2021-04-06
# ℹ 110 more rowsWe can now use complete() to fill in the dates. The complete() function is used like mutate() or summarise(). Unlike these other functions, you need to provide it with a column that exists, and you can’t create new columns. And you also need to provide it with the values that you want completed. In our case, we want each date from the start date to the end date, one for each day. We can use the seq() function to create a sequence of dates, from the min() to the max(). So we’d write it like:
docs/learning.qmd
# A tibble: 823 × 5
id gender age_years intervention date
<dbl> <chr> <dbl> <chr> <date>
1 101 woman 45.6 baseline 2021-03-15
2 101 woman 45.6 baseline 2021-03-16
3 101 woman 45.6 baseline 2021-03-17
4 101 woman 45.6 baseline 2021-03-18
5 101 woman 45.6 baseline 2021-03-19
6 101 woman 45.6 baseline 2021-03-20
7 101 woman 45.6 baseline 2021-03-21
8 101 woman 45.6 baseline 2021-03-22
9 101 woman 45.6 high bioactive diet 2021-05-11
10 101 woman 45.6 high bioactive diet 2021-05-12
# ℹ 813 more rowsWe now have a dataset that we can join with the other datasets! Time to make it into a function 😁
Since we’ve used the tidyr package, let’s add it as a package dependency. Open the Console and run:
Console
usethis::use_package("tidyr")Now, we can make the function. In the docs/learning.qmd file, go to the bottom of the file where we wrote the code to clean the participant details. At the bottom of the chunk, we’ll create a new function named clean_participant_details by assigning function() to it. We’ll only have one argument called data. Then we’ll cut and paste the cleaning code into the function. After that we will replace the participant_details variable with the data argument. Finally, we will assign the cleaned data to a new variable called cleaned and return() it at the end of the function.
Next we’ll go through and add the :: to the functions used, which are tidyr::, tidyselect::, and dplyr::. After that, we will make an Roxygen documentation with Ctrl-Shift-Alt-RCtrl-Shift-Alt-R or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “roxygen comment”).
docs/learning.qmd
#' Convert the participant details data to long and clean it up.
#'
#' @param data The DIME participant details data.
#'
#' @returns A data frame.
#'
clean_participant_details <- function(data) {
cleaned <- data |>
tidyr::pivot_longer(
tidyselect::ends_with("date"),
names_to = NULL, values_to = "date"
) |>
dplyr::group_by(dplyr::pick(-date)) |>
tidyr::complete(
date = seq(min(date), max(date), by = "1 day")
)
return(cleaned)
}Run the function in the code chunk with Ctrl-EnterCtrl-Enter and then test it out by going to the Console and running:
Console
clean_participant_details(participant_details)# A tibble: 823 × 5
id gender age_years intervention date
<dbl> <chr> <dbl> <chr> <date>
1 101 woman 45.6 baseline 2021-03-15
2 101 woman 45.6 baseline 2021-03-16
3 101 woman 45.6 baseline 2021-03-17
4 101 woman 45.6 baseline 2021-03-18
5 101 woman 45.6 baseline 2021-03-19
6 101 woman 45.6 baseline 2021-03-20
7 101 woman 45.6 baseline 2021-03-21
8 101 woman 45.6 baseline 2021-03-22
9 101 woman 45.6 high bioactive diet 2021-05-11
10 101 woman 45.6 high bioactive diet 2021-05-12
# ℹ 813 more rowsNice! It works! Once it works as intended, cut and paste it over into R/functions.R. Then, go to the docs/learning.qmd file and in the setup code chunk, add a pipe to the end of import_dime() for the participant_details object into clean_participant_details(). It should look like this:
docs/learning.qmd
participant_details <- here("data-raw/dime/participant_details.csv") |>
import_dime() |>
clean_participant_details()Then to check the reproducibility, render the document with Ctrl-Shift-KCtrl-Shift-K or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “render”) to make sure it all works. Woohoo 🎉
Like with the pivoting to long section, let them read through this section first and then go over it again to verbally explain it more, making use of the graphs to help illustrate what is happening.
Time: ~10 minutes.
Working in long form can be really useful when doing certain types of wrangling, like you’ve seen and read about above. The majority of the time, you probably will only ever use pivot_longer(). But, sometimes you need to pivot to a form that is a bit wider to do, for example, when doing certain types of joins or processing.
For us, the sleep data is in a very long form, where each hour of sleep has several rows for the different types of sleep. Joining the data in this form to the other datasets, while possible, won’t produce usable output. We want to convert it so there is only one row per hour of a participant’s day, rather than multiple rows per hour. For that, we need to convert to the wider form.
The arguments for pivot_wider() are very similar to those in pivot_longer(), except instead of names_to and values_to, they are called names_from and values_from. Like with many tidyverse functions, the first argument is the data and the other arguments are:
id_cols: This is optional as it will default to all column names. This argument tells pivot_wider() to use the given columns as the identifiers for when converting. Unlike pivot_longer() which doesn’t require some type of “key” or “id” column to convert to long form, the conversion to wide form requires some type of “key” or “id” column because pivot_wider() needs to know which rows belong with each other.names_from: Similar to the pivot_longer(), this is the name of the column that has the values that will make up the new columns. Unlike with the names_to argument in pivot_longer() which takes a character string as input, the column name for names_from must be unquoted, because you are selecting a column that already exists in the dataset (unlike names_to which is the name of a column that you want to create but that doesn’t exist yet).values_from: Same as names_from, this is the column name (that exists and must be given unquoted) for the values that will be in the new columns.Figure 11.3 visually shows what’s happening when using pivot_wider().

N, which is used in the names_from argument, are used as the names for the new columns A and B in the new data.
When you’re ready to continue, place the sticky/paper hat on your computer to indicate this to the teacher 👒 🎩
Time: ~25 minutes.
This will a bit more of a challenging exercise since we haven’t used the pivot_wider() function before. But the way you use it is very similar to pivot_longer(). We’ve had you practice the process many times and this will be an exercise to really challenge what you’ve learned so far! Which is part of the learning journey 😁
For this exercise, you will create a new function that converts the sleep data into a wider form. To test it out, it should look like this and should output something similar:
docs/learning.qmd
sleep_data |>
sleep_types_to_wider()# A tibble: 575 × 10
id date hour seconds_deep seconds_light seconds_wake
<int> <date> <int> <dbl> <dbl> <dbl>
1 101 2021-05-21 23 390 2730 450
2 101 2021-05-22 0 1200 1230 NA
3 101 2021-05-22 1 420 2970 NA
4 101 2021-05-22 2 NA 3870 NA
5 101 2021-05-22 3 NA NA NA
6 101 2021-05-22 4 1710 1200 360
7 101 2021-05-22 5 NA 1680 1350
8 101 2021-05-22 23 330 1710 420
9 101 2021-05-23 0 720 2910 NA
10 101 2021-05-23 1 660 630 570
# ℹ 565 more rows
# ℹ 4 more variables: seconds_rem <dbl>, seconds_asleep <dbl>,
# seconds_restless <dbl>, seconds_awake <dbl>Once you’ve made this new function, move it into the clean_sleep() so it looks like:
R/functions.R
clean_sleep <- function(data) {
cleaned <- data |>
get_participant_id() |>
dplyr::rename(datetime = date) |>
prepare_dates(datetime) |>
# The function used to sum summarise may be different for you.
summarise_column(seconds, list(sum = sum)) |>
# This is the new function you will add below.
sleep_types_to_wider()
return(cleaned)
}Then, you go to your setup code chunk in docs/learning.qmd and run all the code in it, especially:
docs/learning.qmd
sleep_data <- here("data-raw/dime/sleep/") |>
import_csv_files() |>
clean_sleep()It should like this if you run it in the Console:
Console
sleep_data# A tibble: 575 × 10
id date hour seconds_deep seconds_light seconds_wake
<int> <date> <int> <dbl> <dbl> <dbl>
1 101 2021-05-21 23 390 2730 450
2 101 2021-05-22 0 1200 1230 NA
3 101 2021-05-22 1 420 2970 NA
4 101 2021-05-22 2 NA 3870 NA
5 101 2021-05-22 3 NA NA NA
6 101 2021-05-22 4 1710 1200 360
7 101 2021-05-22 5 NA 1680 1350
8 101 2021-05-22 23 330 1710 420
9 101 2021-05-23 0 720 2910 NA
10 101 2021-05-23 1 660 630 570
# ℹ 565 more rows
# ℹ 4 more variables: seconds_rem <dbl>, seconds_asleep <dbl>,
# seconds_restless <dbl>, seconds_awake <dbl>pivot_wider() to get a better idea for how it works.## Pivot wider in the docs/learning.qmd file, then create a new code chunk below it with Ctrl-Alt-ICtrl-Alt-I or with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “new chunk”).sleep_data into pivot_wider(). Try using names_from and values_from to get the right output. You can use the argument names_prefix to add text to the start of the new names columns.After you’ve written some code that works, use the same process you’ve used before to make a function:
sleep_types_to_wider and assign function() to it.data as an argument in function().sleep_data variable with the data argument.wider and return() it at the end of the function.tidyr::.sleep_types_to_wider(sleep_data).R/functions.R.clean_sleep() function in R/functions.R so that it looks like the above.R/functions.R and docs/learning.qmd with the Palette (Ctrl-Shift-PCtrl-Shift-P, then type “style file”).When you’re ready to continue, place the sticky/paper hat on your computer to indicate this to the teacher 👒 🎩
Quickly cover this and get them to do the survey. The previous exercise will likely be challenging for them, so we will not do a discussion activity this time.
pivot_longer() from the tidyr package.pivot_wider().complete() function.This lists some, but not all, of the code used in the section. Some code is incorporated into Markdown content, so is harder to automatically list here in a code chunk. The code below also includes the code from the exercises.
participant_details <- here("data-raw/dime/participant_details.csv") |>
import_dime()
participant_details
participant_details |>
pivot_longer()
participant_details |>
pivot_longer(ends_with("date"))
participant_details |>
pivot_longer(ends_with("date"), names_to = NULL, values_to = "date")
participant_details |>
pivot_longer(ends_with("date"), names_to = NULL, values_to = "date") |>
group_by(pick(-date))
participant_details |>
pivot_longer(ends_with("date"), names_to = NULL, values_to = "date") |>
group_by(pick(-date)) |>
complete(
date = seq(min(date), max(date), by = "1 day")
)
usethis::use_package("tidyr")
#' Convert the participant details data to long and clean it up.
#'
#' @param data The DIME participant details data.
#'
#' @returns A data frame.
#'
clean_participant_details <- function(data) {
cleaned <- data |>
tidyr::pivot_longer(
tidyselect::ends_with("date"),
names_to = NULL, values_to = "date"
) |>
dplyr::group_by(dplyr::pick(-date)) |>
tidyr::complete(
date = seq(min(date), max(date), by = "1 day")
)
return(cleaned)
}
clean_participant_details(participant_details)
participant_details <- here("data-raw/dime/participant_details.csv") |>
import_dime() |>
clean_participant_details()
#' Convert the sleep types to wide format.
#'
#' @param data The cleaned DIME sleep data.
#'
#' @returns A data frame.
#'
sleep_types_to_wider <- function(data) {
wider <- data |>
tidyr::pivot_wider(
names_from = sleep_type,
names_prefix = "seconds_",
values_from = seconds_sum
)
return(wider)
}